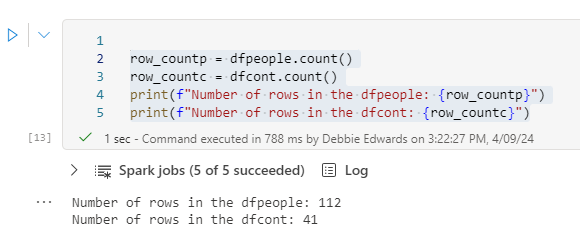
Now we have updated our ProcessedFiles delta parquet, we can update the dimensions accordingly.
- It might be nice to have another delta parquet file here we can use to collect meta data for dims and facts.
- We will also want the processedDate in each dim and fact table.
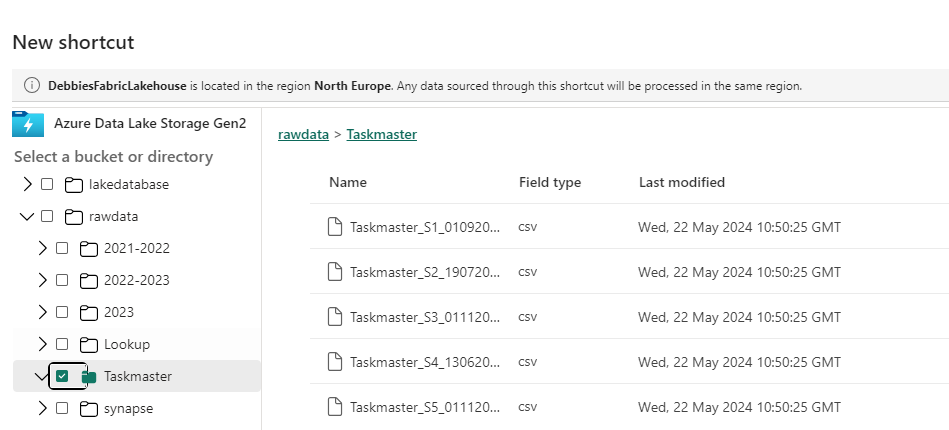
We are going to start with Delta Load 2. So. Series 1 and 2 have already been loaded. We are now loading Series 3 (This shows off some of the logic better)
Dim Contestant.
Bring back the list of Processed Files from ProcessedFiles
Immediately we have a change to the code because its delta Parquet and not Parquet
# Define the ABFS path to the Delta table
delta_table_path = "abfss://########-####-####-####-############@onelake.dfs.fabric.microsoft.com/########-####-####-####-############/Tables/processedfiles"
# Read the Delta table
dflog = spark.read.format("delta").load(delta_table_path)
# Filter the DataFrame
df_currentProcess = dflog[dflog["fullyProcessedFlag"] == 0][["Filename"]]
# Display the DataFrame
display(df_currentProcess)

New Code Block Check processed Files
In the previous code, we:
- Create a new taskmaster schema to load data into
- Loop through the files and add data to the above schema
- Add the current process data into CurrentProcessedTMData.parquet.
- Add the ProcessDate of the Taskmaster transformed data
We know at some point that we are having issues with the fact table getting duplicate series so we need to check throughout the process where this could be happening.
- Change the processDate from being the TaskmasterTransformed process date, in case this was done on an earlier day. We want the current date here to ensure everything groups together.
- We could also set up this data so we can store it in our new Delta parquet Log
- Because we are also adding process date to the group. we need to be careful. minutes and seconds could create extra rows and we don’t want that
- We also know that we are doing checks on the data as it goes in. And rechecking the numbers of historical data. so we need a flag for this
from pyspark.sql import functions as F
workspace_id = "########-####-####-####-############"
lakehouse_id = "########-####-####-####-############"
# Define the file path
file_path = f"abfss://{workspace_id}@onelake.dfs.fabric.microsoft.com/{lakehouse_id}/Files/Data/Silver/Log/CurrentProcessedTMData.parquet"
# Read the Parquet file
dfCheckCurrentProc = spark.read.parquet(file_path)
# Add the new column
dfCheckCurrentProc = dfCheckCurrentProc.withColumn("sequence", F.lit(1))
dfCheckCurrentProc = dfCheckCurrentProc.withColumn("parquetType", F.lit("parquet"))
dfCheckCurrentProc = dfCheckCurrentProc.withColumn("analyticsType", F.lit("Transformation"))
dfCheckCurrentProc = dfCheckCurrentProc.withColumn("name", F.lit("CurrentProcessedTMData"))
# Set the date as the current date
dfCheckCurrentProc = dfCheckCurrentProc.withColumn("processedDate", current_date())
dfCheckCurrentProc = dfCheckCurrentProc.withColumn("historicalCheckFlag", F.lit(0))
dfCheckCurrentProc = dfCheckCurrentProc.withColumn("raiseerrorFlag", F.lit(0))
# Select the required columns and count the rows
dfCheckCurrentProc = dfCheckCurrentProc.groupBy("sequence","parquetType","analyticsType","name","source_filename", "processedDate","historicalCheckFlag","raiseerrorFlag").count()
# Rename the count column to TotalRows
dfCheckCurrentProc = dfCheckCurrentProc.withColumnRenamed("count", "totalRows")
# Show the data
display(dfCheckCurrentProc)- current_date() gets the current date. We only have date in the grouped auditing table. date and time can go into the dimension itsself in the process date.
- sequence allows us to see the sequence of dim and fact creation
- parquetType is either Parquet or Delta Parquet for this project
- analyticsType is the type of table we are dealing with
- historicalCheckFlag. Set to 0 if we are looking at the data being loaded. 1 if the checks are against older data.
- raiseErrorflag if there looks like we have any issues. This can be set to 1 and someone can be alerted using power BI Reporting.
It would be really good to also have a run number here because each run will consist of a number of parquet files. In this case parquet and delta parquet

Get ContestantTransformed data
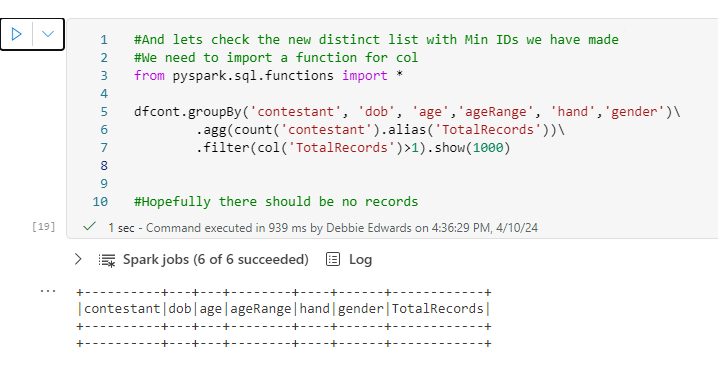
A small update here. Just in case, we use distinct to make sure we only have distinct contestants in this list
# Ensure the DataFrame contains only distinct values
dfc = dfc.distinct()
Merge Contestants and Taskmaster data
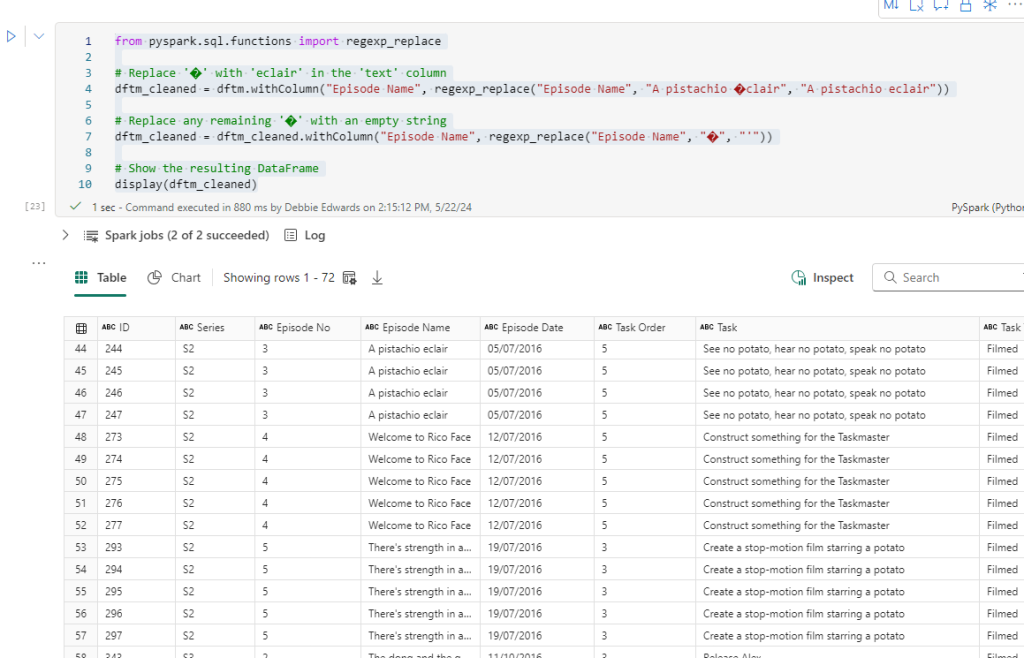
This is the point where we create the Contestant Dimension. the processDate to the current date and alias to to just processedTime.
from pyspark.sql.functions import col, current_date
from pyspark.sql import SparkSession, functions as F
df_small = F.broadcast(dfc)
# Perform the left join
merged_df = dftm.join(df_small, dftm["Contestant"] == df_small["Name"], "left_outer")\
.select(
dfc["Contestant ID"],
dftm["Contestant"].alias("Contestant Name"),
dftm["Team"],
dfc["Image"],
dfc["From"],
dfc["Area"],
dfc["country"].alias("Country"),
dfc["seat"].alias("Seat"),
dfc["gender"].alias("Gender"),
dfc["hand"].alias("Hand"),
dfc["age"].alias("Age"),
dfc["ageRange"].alias("Age Range"),
current_timestamp().alias("processedTime"),
).distinct()
# Show the resulting DataFrame
merged_df.show()
Add the Default row
This changes because we have a new column that we need to add.
from pyspark.sql import SparkSession,Row
from pyspark.conf import SparkConf
from pyspark.sql.functions import current_timestamp
workspace_id = "########-####-####-####-############"
lakehouse_id = "########-####-####-####-############"
path_to_parquet_file = (f"abfss://{workspace_id}@onelake.dfs.fabric.microsoft.com/{lakehouse_id}/Tables/dimcontestant")
# Check if the Delta Parquet file exists
delta_file_exists = spark._jvm.org.apache.hadoop.fs.FileSystem.get(spark._jsc.hadoopConfiguration()).exists(spark._jvm.org.apache.hadoop.fs.Path(path_to_parquet_file))
if delta_file_exists:
print("Delta Parquet file exists. We don't need to add the NA row")
else:
# Add a Default row
# Create a sample DataFrame
data = [(-1, "Not Known","Not Known","Not Known","Not Known","Not Known","Not Known","0","NA","0","0","0")]
columns = ["Contestant ID", "Contestant Name", "Team","Image","From","Area","Country","Seat","Gender","Hand","Age","Age Range"]
new_row = spark.createDataFrame(data, columns)
# Add the current timestamp to the ProcessedTime column
new_row = new_row.withColumn("ProcessedTime", current_timestamp())
# Union the new row with the existing DataFrame
dfContfinal = dfContfinal.union(new_row)
# Show the updated DataFrame
dfContfinal.show(1000)
print("Delta Parquet file does not exist. Add NA Row")
The above screen show is for Series 1 and 2. Just to show you that the row is created if its the first time.
All the process dates are the same (at least the same day) ensuring everything will be grouped when we add it to the audit delta parquet table.

However, this is displayed for our 2nd load for series 3 (2nd load)
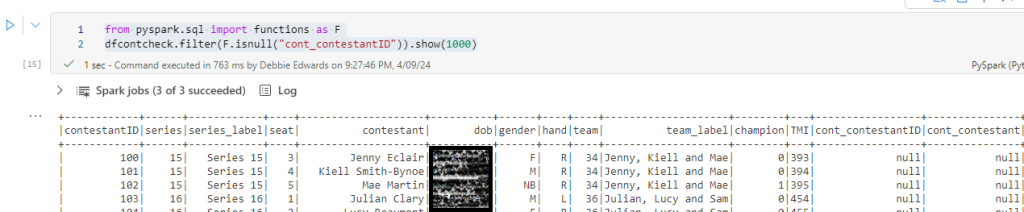
Check the new data that has been loaded into the delta parquet.
Checking the data, checks the dimension that has been created as delta parquet This is the last code section and we want to build more in after this

Get the Contestant Names and Source Files of the Current Load
Because we can process either 1 file alone, or multiple files. we need to get the source file name AND the contestant name at this point.
#Get just the contestant name and source File name from our currrent load
df_contestant_name = dftm.selectExpr(“`Contestant` as contestantNameFinal”, “`source_filename` as sourceFileNameFinal”).distinct()
df_contestant_name.show()

For the audit we need to know what our current Contestants are and who are the Contestants we have previously worked with. Also the original file they below too.
We took this information from an earlier dataframe when we loaded the source file to be used to create the dimension. This still contains the source file name. The Dimension does not contain the file name any more.
Join df_contestant_name to dfnewdimc to see what data is new and what data has been previously loaded
from pyspark.sql.functions import broadcast, when, col
df_joined = dfnewdimc.join(
df_contestant_name,
dfnewdimc.ContestantName == df_contestant_name.contestantNameFinal,
how='left'
)
# Add the currentFlag column
df_joined2 = df_joined.withColumn(
"historicalCheckFlag",
when((col("contestantNameFinal").isNull()) & (col("ContestantKey") != -1), 1).otherwise(0))
# Drop the ContestantNameFinal column
df_joined3 = df_joined2.drop("contestantNameFinal")
#If its an audit row set sourceFileNameFinal to Calculated Row
df_joined4 = df_joined3.withColumn(
"sourceFileNameFinal",
when((col("sourceFileNameFinal").isNull()) & (col("ContestantKey") == -1), "Calculated Row")
.otherwise(col("sourceFileNameFinal"))
)
# Show the result
display(df_joined4)
This joins the name we have just created to the data we are working with so we can set the historicalCheckFlag. For the first load everything is 0 because they are current.
In this load we can see some 1’s These are records already in the system and don’t match to our current load.
This file also now contains the source_filename
If its a default row, there will be no source filename so we set to calculated Row instead
Creating Dim Audit row
from pyspark.sql import functions as F
from pyspark.sql import SparkSession
from pyspark.sql.types import StructType, StructField, StringType, IntegerType, TimestampType
from pyspark.sql import DataFrame
# Add the new column
dfdimaudit = df_joined4.withColumn("sequence", F.lit(2))
dfdimaudit = dfdimaudit.withColumn("parquetType", F.lit("Delta parquet"))
dfdimaudit = dfdimaudit.withColumn("analyticsType", F.lit("Dimension"))
dfdimaudit = dfdimaudit.withColumn("name", F.lit("Dim Contestant"))
# Extract the date from source_processedTime
dfdimaudit = dfdimaudit.withColumn("processedDate", F.to_date("processedTime"))
dfdimaudit = dfdimaudit.withColumn("raiseerrorFlag", F.lit(0))
# Select the required columns and count the rows
dfdimauditgr = dfdimaudit.groupBy("sequence","parquetType","analyticsType","name", "sourceFileNameFinal","processedDate","historicalCheckFlag","raiseerrorFlag").count()
# Alias sourceFileNameFinal to sourceFileName
dfdimauditgr = dfdimauditgr.withColumnRenamed("sourceFileNameFinal", "source_filename")
# Alias count to totalRowsdfdimauditgr = dfdimauditgr.withColumnRenamed("count", "totalRows")
# Show the data
display(dfdimauditgr) The files are grouped on processed time. So we need to deal with this accordingly so set date and time to just date using to_date() (To avoid minutes and seconds causing issues)

Our default row is set against, calculated Row.
The null source file name is the total count of our historical records
We are going to append the dim audit to the initial current processed audit so they both need exactly the same columns and these are
- sequence
- parquetType
- analyticsType
- name
- source_filename
- processedDate
- totalRows
- historicalCheckFlag
- raiseerrorFlag
and we have the correct source file name. Even if we process multiple files.
Add Historical Data Check to null source filename

Append the audit for the transformation data and Dimension data
# Append the current processed transformation file(s) audit row with the dimension audit
audit3_df = dfCheckCurrentProc.unionByName(dfdimauditgr)
display(audit3_df) 
With the S3 file loaded we can see
- Transformation rows for series 3.
- Dimension rows for Contestant S3
- The historical row count of all series already in the dimension.
Create Batch numbers for the audit.
If its the first time we run this process. Set to 0. Else set to the Match Last Batch No + 1
from pyspark.sql.functions import lit
# Function to check if a table exists
def table_exists(spark, table_name):
return spark._jsparkSession.catalog().tableExists(table_name)
# Table name
table_name = "SilverDebbiesTraininglh.processeddimfact"
if table_exists(spark, table_name):
# If the table exists, get the max batchno
df = spark.table(table_name)
max_batchno = df.agg({"batchno": "max"}).collect()[0][0]
next_batchno = max_batchno+1
# Add the batchNo column with a value of 0
audit3_df = audit3_df.withColumn("batchNo", lit(next_batchno))
print(f"processedDimFact data already exists. Next batchno: {next_batchno}")
display(audit3_df)
else:
# Add the batchNo column with a value of 0
audit3_df = audit3_df.withColumn("batchNo", lit(0))
# Reorder columns to make batchNo the first column
columns = ['batchNo'] + [col for col in audit3_df.columns if col != 'batchNo']
audit3_df = audit3_df.select(columns)
print("Table does not exist. Add Batch No 0")
display(audit3_df)

The following gives more information about the code used.

def table_exists(spark, table_name):defines a function named table_exsts that takes two parameters. the spark session object and the table name- Next we define a function named table_exists that will look for the table name
spark._jsparkSession.catalog()accesses the spark session catalogue which contains metadata about the entities in the spark session.- Basically, this is checking if the table name exists in the spark session catalogue. It it does we get the match Batch no from the table. Add 1 and add that as the column next_batchno
- If its false we start batchno at 0
How does the column reordering work?

- [‘batchNo’]: This creates a list with a single element, ‘batchNo’. This will be the first column in the new order.
- [col for col in audit3_df.columns if col != ‘batchNo’]: This is a list comprehension that iterates over all the columns in audit3_df and includes each column name in the new list, if removes ‘batchNo’ from the list.
List Comprehension: Create lists by processing existing lists (Like columns here) into a single line of code.
- Combining the lists: The + operator concatenates the two lists. So, [‘batchNo’] is combined with the list of all other columns, resulting in a new list where ‘batchNo’ is the first element, followed by all the other columns in their original order.

And here we can see in Load 2 we hit the if section of the above code. Our batch no is 1 (Because the first load was 0)
Reset totalRows to Integer

Found that totalRows was set to long again so before adding into delta parquet. reset to int.
Bring back the processeddimFact before saving with the new rows
Before saving the new audit rows we want to know if there are any issues. This means we need the data. However we also need to deal with the fact that this may be the first process time and there won’t be a file to check.
table_path = "abfss://########-####-####-####-############@onelake.dfs.fabric.microsoft.com/########-####-####-####-############/Tables/processeddimfact"
table_exists = DeltaTable.isDeltaTable(spark, table_path)
if table_exists:
# Load the Delta table as a DataFrame
dfprocesseddimfact = spark.read.format("delta").table("SilverDebbiesTraininglh.processeddimfact")
# Display the table
dfprocesseddimfact.show() # Use show() instead of display() in standard PySpark
# Print the schema
dfprocesseddimfact.printSchema()
print("The Delta table SilverDebbiesTraininglh.processeddimfact exists. Bring back data")
else:
# Define the schema
schema = StructType([
StructField("batchNo", IntegerType(), nullable=False),
StructField("sequence", IntegerType(), nullable=False),
StructField("parquetType", StringType(), nullable=False),
StructField("analyticsType", StringType(), nullable=False),
StructField("name", StringType(), nullable=False),
StructField("source_filename", StringType(), nullable=True),
StructField("processedDate", DateType(), nullable=True),
StructField("historicalCheckFlag", IntegerType(), nullable=False),
StructField("raiseerrorFlag", IntegerType(), nullable=False),
StructField("totalRows", IntegerType(), nullable=False)
])
# Create an empty DataFrame with the schema
dfprocesseddimfact = spark.createDataFrame([], schema)
dfprocesseddimfact.show()
print("The Delta table SilverDebbiesTraininglh.processeddimfact does not exist. Create empty dataframe")

If there is no data we can create an empty schema.
Get the Total of Dim Contestant rows (None Historical)
from pyspark.sql.functions import col, sum
# Filter the dataframe where name is "Dim Contestant"
filtered_df = dfprocesseddimfact.filter((col("Name") == "Dim Contestant") & (col("historicalCheckFlag") == 0)& (col("source_filename") != "Calculated Row"))
display(filtered_df)
# Calculate the sum of TotalRows
total_rows_sum = filtered_df.groupBy("Name").agg(sum("TotalRows").alias("TotalRowsSum"))
# Rename the "Name" column to "SelectedName"
total_rows_sum = total_rows_sum.withColumnRenamed("Name", "SelectedName")
display(total_rows_sum)
# Print the result
print(f"The sum of TotalRows where name is 'Dim Contestant' is: {total_rows_sum}")
Both Initial (None Historical Loads) equal 10
Get Historical Rows from our latest process and join to total Loads of the previous Load
#Join total_rows_sum to audit3_df
from pyspark.sql.functions import broadcast
# Broadcast the total_rows_sum dataframe
broadcast_total_rows_sum = broadcast(total_rows_sum)
# Perform the left join
df_checkolddata = audit3_df.filter(
(audit3_df["historicalCheckFlag"] == 1) & (audit3_df["source_filename"] != "Calculated Row")
).join(
broadcast_total_rows_sum,
audit3_df["name"] == broadcast_total_rows_sum["SelectedName"],
"left"
)
# Show the result
display(df_checkolddata)
So Series 1 and 2 have 10 contestants. After S3 load there are still 10 contestants. Everything is fine here. The historical records row allows us to ensure we haven’t caused any issues.
Set Raise Error Flag if there are issues
And now we can set the error flag if there is a problem which can raise an alert.
from pyspark.sql.functions import col
# Assuming df is your DataFrame
total_rows_sum = df_checkolddata.agg({"TotalRowsSum": "sum"}).collect()[0][0]
total_rows = df_checkolddata.agg({"totalRows": "sum"}).collect()[0][0]
# Check if TotalRowsSum equals totalRows
if total_rows_sum == total_rows:
print("No issue with data")
else:
df_checkolddata = df_checkolddata.withColumn("raiseerrorFlag", lit(1))
print("Old and historical check does not match. Issue. Set raiseerrorFlag = 1")
# Show the updated DataFrame
df_checkolddata.show()
raiseerrorFlag is still 0
Remove the historical check row if there is no error
from pyspark.sql.functions import col
# Alias the DataFrames
audit3_df_alias = audit3_df.alias("a")
df_checkolddata_alias = df_checkolddata.alias("b")
# Perform left join on the name column and historicalCheckFlag
df_audit4 = audit3_df_alias.join(df_checkolddata_alias,
(col("a.name") == col("b.name")) &
(col("a.historicalCheckFlag") == col("b.historicalCheckFlag")),
how="left")
# Select only the columns from audit3_df and TotalRowsSum from df_checkolddata, and alias historicalCheckFlag
df_audit5 = df_audit4.select(
"a.*",
col("b.raiseerrorFlag").alias("newraiseErrorFlag")
)
# Filter out rows where newraiseerrorflag is 0 because its fine. We don';'t need to therefore know about the historical update
df_audit6 = df_audit5.filter((col("newraiseErrorFlag") != 0) | (col("newraiseErrorFlag").isNull()))
# Remove the columns newraiseerrorflag
df_audit7 = df_audit6.drop("newraiseErrorflag")
# Show the result
display(df_audit7)
the historical row has been removed. if there isn’t a problem, there is no need to hold this record.
If the data has multiple audit rows. Flag as an issue. Else remove
# Count the number of rows where ContestantName is "Not Known"
dfcount_not_known = dfnewdimc.filter(col("ContestantName") == "Not Known").count()
# Print the result
print(f"Number of rows where ContestantName is 'Not Known': {dfcount_not_known}")
# Check if count_not_known is 1
if dfcount_not_known == 1:
# Remove row from df_audit7 where source_filename is "CalculatedRow"
df_audit8 = df_audit7.filter(col("source_filename") != "Calculated Row")
print(f"1 dim audit row. We can remove the row from the audit table")
else:
# Set source_filename to "Multiple Calculated Rows issue"
df_audit8 = df_audit7.withColumn("source_filename",
when(col("source_filename") == "Calculated Row",
lit("Multiple Calculated Rows issue"))
.otherwise(col("source_filename")))
print(f"multiple dim audit row. We can specify this in the audit table")
# Show the updated df_audit7
df_audit8.show()
The Audit rows has now been removed. Unless there is an issue we don’t need it. this will be useful if we accidentally process multiple audit rows into the table.
Save as Delta Parquet
from delta.tables import DeltaTable
#You can also add the none default daya lake by clicking +Lakehouse
audit3_df.write.mode("append").option("overwriteSchema", "true").format("delta").saveAsTable("SilverDebbiesTraininglh.processedDimFact")
# Print a success message
print("Parquet file appended successfully.")
And save the data as a Delta Parquet processedDimFact in our Silver transform layer.
We can use both of these Delta Parquet Files later.
Check processdimfact

Dim Contestant is complete for now. But we will come back later to update the code.
Dim Episode
Create Episode Dim
Small change. We are adding processedTime as the current date and time.
from pyspark.sql.functions import current_timestamp
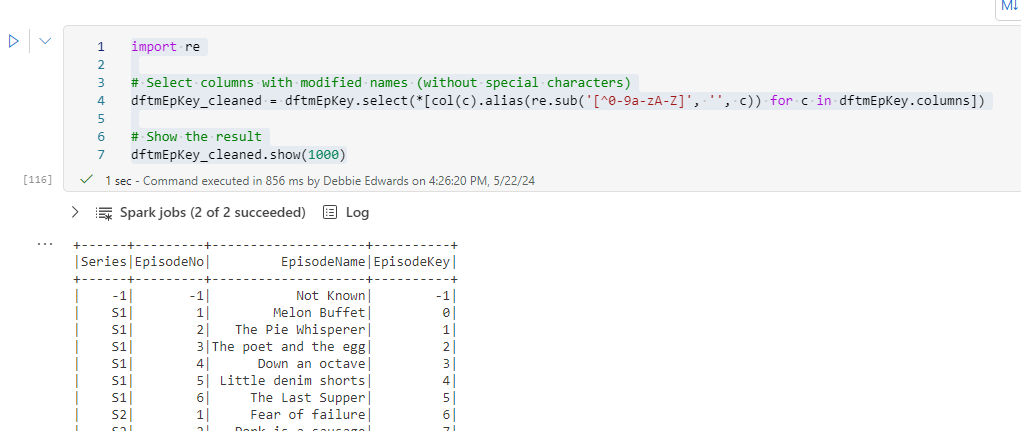
dftmEp = dftm.select("Series","Episode No" ,"Episode Name").distinct()
# Add the current timestamp as processedTime
dftmEp = dftmEp.withColumn("processedTime", current_timestamp())
dftmEp = dftmEp.orderBy("Series", "Episode No")
display(dftmEp)
Add a Default row
Just like Contestant. We need ProcessedTime adding
from pyspark.sql.functions import current_timestamp # Add the current timestamp to the ProcessedTime column
new_row = new_row.withColumn("ProcessedTime", current_timestamp())


Again, Here is the screenshot taken on the first load of S1 and S2

And our S3 Load 2 hits the if block and no default row is created
Get the Series, Episode Name and Source File of the Processed data
#Get just the series, Episode Name and source_filename from our current load
df_SeEp = dftm.selectExpr("`Series` as SeriesFinal", "`Episode Name` as EpisodeFinal", "source_filename").distinct()
display(df_SeEp) 
` are used in PySpark to handle column names that contain special characters, spaces, or reserved keywords.
Join df_SeEp to dfnewdimt
Join the list pf currently processed series and episodes to everything in the dimension so far (the data frame we created when we checked the dim
from pyspark.sql.functions import broadcast, when, col
# Broadcast df_contestant_name and perform the left join
df_joined = dfnewdimt.join(
broadcast(df_SeEp),
(dfnewdimt.Series == df_SeEp.SeriesFinal) & (dfnewdimt.EpisodeName == df_SeEp.EpisodeFinal),
"left"
)
# Add the currentFlag column
df_joined2 = df_joined.withColumn("historicalCheckFlag", when(col("SeriesFinal").isNull(), 1).otherwise(0))
# Drop the ContestantNameFinal column
df_joined3 = df_joined2.drop("SeriesFinal").drop("EpisodeFinal")
#If its an audit row set sourceFileNameFinal to Calculated Row
df_joined4 = df_joined3.withColumn(
"source_filename",
when((col("source_filename").isNull()) & (col("EpisodeKey") == -1), "Calculated Row")
.otherwise(col("source_filename"))
)
# Show the result
display(df_joined4)
All historicalCheckFlags are set to 0 if its the current Load (s3). 1 when there is no match and its historical data. the source_filename from df.SeEp has been left in the result which can be used later.
Again we have set the Episode Keys source_filename to “calculated Row” because there is no source to join this one record on by the series and episode name.
Add audit row
At the end of the process. We will add an audit row
from pyspark.sql import functions as F
# Add the new column
dfdimaudit = df_joined4.withColumn("sequence", F.lit(3))
dfdimaudit = dfdimaudit.withColumn("parquetType", F.lit("Delta parquet"))
dfdimaudit = dfdimaudit.withColumn("analyticsType", F.lit("Dimension"))
dfdimaudit = dfdimaudit.withColumn("name", F.lit("Dim Episode"))
# Extract the date from source_processedTime
dfdimaudit = dfdimaudit.withColumn("processedDate", F.to_date("processedTime"))
dfdimaudit = dfdimaudit.withColumn("raiseerrorFlag", F.lit(0))
# Select the required columns and count the rows
dfdimauditgr = dfdimaudit.groupBy("sequence","parquetType","analyticsType","name", "source_filename","processedDate","historicalCheckFlag","raiseerrorFlag").count()
# Alias count to totalRows
dfdimauditgr = dfdimauditgr.withColumnRenamed("count", "totalRows")
# Show the data
display(dfdimauditgr) 
for Load 2 we have 5 records in the current load with the source filename set. The historical load of 11 and the calculated Audit -1 row.
Bring back our processeddimfact data
from delta.tables import DeltaTable
from pyspark.sql.functions import col, max
# Load the Delta table as a DataFrame
dfprocesseddimfact = spark.read.format("delta").table("SilverDebbiesTraininglh.processeddimfact")
# Find the maximum BatchNo
max_batch_no = dfprocesseddimfact.agg(max("BatchNo")).collect()[0][0]
# Filter the DataFrame to include only rows with the maximum BatchNo
dfprocesseddimfact = dfprocesseddimfact.filter(col("BatchNo") == max_batch_no)
# Display the table
display(dfprocesseddimfact)
dfprocesseddimfact.printSchema()
Here we find the max batch number and then filter our data frame to only give us the max batch number which is the one we are working on. (Because at this point, we are in the middle of a batch of dims and facts)
Create a distinct batch No
distinct_batch_no = dfprocesseddimfact.select("batchNo").distinct()
display(distinct_batch_no)

We just want that distinct Max Batch number for the next process
Join to our Dim Episode Audit row to bring in the batch number via Left outer join
# Alias the dataframes
dfdimauditgr_alias = dfdimauditgr.alias("dfdimauditgr")
distinct_batch_no_alias = distinct_batch_no.alias("dfprocesseddimfact")
# Perform inner join
audit2_df = dfdimauditgr_alias.crossJoin(distinct_batch_no_alias)
# Select specific columns
audit2_df = audit2_df.select(
distinct_batch_no_alias.batchNo,
dfdimauditgr_alias.sequence,
dfdimauditgr_alias.parquetType,
dfdimauditgr_alias.analyticsType,
dfdimauditgr_alias.name,
dfdimauditgr_alias.source_filename,
dfdimauditgr_alias.totalRows,
dfdimauditgr_alias.processedDate,
dfdimauditgr_alias.historicalCheckFlag,
dfdimauditgr_alias.raiseerrorFlag
)
audit2_df = audit2_df.distinct()
# Show result
display(audit2_df)
The cross join (Cartesian Join) is because there is only 1 batch number set in distinct_batch_no (the Max Batch number is the one we are working on) So no need to add join criteria.
Set totalRows to Int

Once again TotalRows needs setting to Int
Get the processeddimfact data
# Load the Delta table as a DataFrame
dfprocesseddimfact = spark.read.format("delta").table("SilverDebbiesTraininglh.processeddimfact")
# Display the table
dfprocesseddimfact.show() # Use show() instead of display() in standard PySpark
# Print the schema
dfprocesseddimfact.printSchema() # Load the Delta table as a DataFrame
dfprocesseddimfact = spark.read.format("delta").table("SilverDebbiesTraininglh.processeddimfact")
# Display the table
dfprocesseddimfact.show() # Use show() instead of display() in standard PySpark
# Print the schema
dfprocesseddimfact.printSchema()
We will need this to check our historical checked data against the original update
Get the total rows of our None historical rows that have been previously loaded

Same logic as Contestant
Join Historical records with new audit to see if the numbers have changed

No changes to the old numbers.
Set raiseerrorFlag if there are issues

There are no issues
If no issues. Record is removed

As there are no issues the record is removed
If Audit row -1 is only in once. No need to hold the audit row

Append into processeddimfact

And check the data looks good

We head off into Dim Task now and here is where we hit some issues. We need to add more code into the process.
Dim Task
checking the count of rows between the original load, and the historical check

When we count the rows of processed data they come to 64. But when we run the check again it comes to 65. Which would trigger the raise error flag. And this is why
dfnewdimt_filtered = dfnewdimt.filter(dfnewdimt["Task"] == "Buy a gift for the Taskmaster")
display(dfnewdimt_filtered)
I settled on Task and TaskOrder to create the unique row. However this isn’t the case. we have two series where our Task and Task Order are identical.
What we really want is one record in the dimension applied to the correct seasons in the fact table. Not a duplicate like we have above.
The following code is adding Series 3 to both “Buy a gift for the taskmaster” and is consequently undercounting.
Series three is a good opportunity to update the logic as we know there is a record already added from season 2
Bring back old dimTask
from delta.tables import DeltaTable
from pyspark.sql import SparkSession
from pyspark.sql.types import StructType, StructField, StringType, IntegerType, TimestampType
# Initialize Spark session
#spark = SparkSession.builder.appName("DeltaTableCheck").getOrCreate()
# Path to the Delta table
table_path = "abfss://########-####-####-####-############@onelake.dfs.fabric.microsoft.com/########-####-####-####-############/Tables/dimtask"
# Check if the Delta table exists
if DeltaTable.isDeltaTable(spark, table_path):
# Load the Delta table as a DataFrame
dfprevdimt = spark.read.format("delta").load(table_path)
else:
# Define the schema for the empty DataFrame
schema = StructType([
StructField("Task", StringType(), True),
StructField("TaskOrder", IntegerType(), True),
StructField("TaskType", StringType(), True),
StructField("Assignment", StringType(), True),
StructField("ProcessedTime", TimestampType(), True)
])
# Create an empty DataFrame with the specified schema
dfprevdimt = spark.createDataFrame([], schema)
print("no dim. Create empty dataframe")
# Display the DataFrame
dfprevdimt.show()
If its the first time, create an empty dataset. else bring back the dataset
Join new Tasks to all the Previously added tasks

The decision has been made to use all the columns in the join, Just in case a Task has a different assignment or Type.
Our Task that we know is identical for Series 2 and 3 appears. We use the dftm dataframe. This contains the data before we create the distinct dimension, as this contains source_filename. Inner Join only brings back the matching records.
Add this to a Delta Parquet File


We can now have a static item we can use later. and we can report on these items
If task already exists. Remove from the new load
from pyspark.sql.functions import col,current_timestamp
# Alias the DataFrames
dfprevdimt_df_alias = dfprevdimt.alias("old")
dftm_alias = dftm.alias("new")
# Perform left join on the name column and historicalCheckFlag
dftmdupRemoved = dftm_alias.join(dfprevdimt_df_alias,
(col("new.Task") == col("old.Task")) &
(col("new.Task Order") == col("old.TaskOrder"))&
(col("new.Task Type") == col("old.TaskType"))&
(col("new.Assignment") == col("old.Assignment")),
how="Left_anti")
#And reset the dimension
dfTasksdupRemoved = dftmdupRemoved.select("Task","Task Order" ,"Task Type", "Assignment").distinct()
# Add the current timestamp as processedTime
dfTasksdupRemoved = dfTasksdupRemoved.withColumn("processedTime", current_timestamp())
# Show the result
display(dfTasksdupRemoved)
The left anti join bring back records that exist in the new data frame and removes anything that also exists in the pre existing data
dtfm is again used so we have to reset the dimension.
dfTasksdupRemoved now becomes our main dataframe so we need to ensure its used across the Notebook where neccessary
We then go on to create the default row (If its never been run before) and keys
Add a Default Row
Code changed to use dfTasksdupRemoved

Create the Task Key
Again, we have had a change to code to use the updated dataframe


Get Task details from current load

Previously, This was just Task and Order but we know that we need more in the join.
Create a historical Record check



Buy a gift for the Taskmaster is flagged as historical (Its in Series 2) and we have removed it from series 3. And we have added this information to a Delta Parquet.
We have also added more join criteria.
Create the Audit Rows

Add a value into source filename if full because its a historical record

Bring back the ProcessedDimfact

Get the Current batch

Join the batch number into the dataframe

Load the processed Dim Fact

Count the rows
Total Rows is correct at 65

Add the Rows of the historical check against rows that were originally processed

These now match
If Issue. Set RaiseErrorFlag = 1

No issues
Remove historical check if there are no issues

Check that there aren’t multiple rows in the data for the default -1 row. If all ok. remove default row from audit

And append the data into processeddimfact.
There is other logic to consider here. Imagine if we loaded in Series 1,2 and 3 together. Would the logic still remove the duplicate task. This will need running again to test
Taskmaster Fact
The fact table doesn’t have default records so we need to deal with this slightly differently. Lets run through the code again
The new fact data has been processed at the point we continue with the logic
Get Data of current process we can join on

Join current and already processed Fact data
- df_currentfact is the current S3 data
- dfnewfact is the newly loaded fact table that contains everything. S1,2 and 3.
dfnewfact is on the left of the join. So logically. If current and new both exist we get the match.

Create the Audit Row

If source Filename is null. Add Historical Data Check

Bring back the latest batch from processeddimFact

Get Current Batch No

Add the batch number to the audit

Load all of the processed dim fact

Total Up the none historical rows (totals created at the time)

Add the total rows (As at the time they were created to the total historical rows)

We can already see they match. 307 rows for S1 and S2
Reset the Raise Error flag to 1 if there is a problem

Remove the historical check row if there are no issues

And now you can append into the Delta parquet table and check the processed dim and fact.
And then we are back to setting the processedflag to 1 at the end of the fact table.
Conclusion
Now we have an audit of our Loads (2 Loads so far)

We have done a lot of work creating auditing. We can now add files and hopefully run the pipeline to automate the run. then we can create Power BI reporting.
We now have three audit Delta parquet tables
- processedFiles – This goes through each Taskmaster Series File and allows you to see what you are working on. the fullyProcessedFlag is set to 0 when the fact table is complete
- processeddimFact – (Shown above) The detailed audit for the transformation, dim and fact tables. Containing, number of records. The date time processed. And a flag for issues
- dimremovenoneuniquetaskaudit. Tasks are possible to duplicate. For example, we have the same task (And all of its components like sequence) are identical between series 2 and 3. We only need the one row so the duplicate is removed
There is more that we could do with this. of course there always is. However the big one is to make sure that we get none duplicated task information if series 2 and 3 are processed together.
We could also add more detail into the audit. For example any issues with the data such as Column A must always be populated with either A B C or D. If Null then flag as error. If anything other that specified codes then error.
We will come back to this but in the next blog I want to try something a bit different and set up the Data Warehouse. How would this project look if using the warehouse. And why would you choose one over the other.
So in part 19 we will look at setting the warehouse up and how we deal with the source data in the data lake